Whole genome sequencing
An Exeter senior maps the fruit fly genome. Along the way he finds new mysteries to explore.
At the end of winter term, students in BIO 586: Molecular Genetics prep fruit fly DNA for the first whole genome sequence ever created at Exeter.
It’s 10 a.m. on Sunday, a few days before the end of winter term, and 25 students are gathering in classroom 307 of Phelps Science Center to do something that no one at Exeter has done before. As cool light pours in through the windows, the students unbundle from coats and sweatshirts. Twelve sit at the Harkness table in the inside corner of the room. The others perch on high stools or lean against lab tables. Dotted throughout are indicators of the room’s focus: beakers, plastic models of chromosomes, 12 microscopes, a near-life-size human anatomical model. The students, enrolled in two sections of BIO 586: Molecular Genetics, are here on Sunday because it’s the one day that allows for extended unscheduled time. They have the relaxed poise of teenagers unhooked from weekday or Saturday demands.
Today’s task is the “wet lab” prep work that will isolate, clean and amplify (or copy) small snippets of DNA from four lines of Drosophila melanogaster, the common fruit fly. These snippets will be packed with dry ice and shipped to a lab for sequencing. The hope is that the resulting data will be accurate enough to allow an Exeter senior, Sanath Govindarajan, to analyze it and assemble a whole genome for one or more of the lines as a spring-term project. A lot is riding on today.
All eyes are on Science Instructor Anne Rankin ’92 as she describes the protocol. There will be four procedures, she explains. You will work in pairs at your own pace, but we will wait until everyone is done to start the next procedure. Avoiding contamination is essential, so wear gloves and use new pipette tips at each step. “We will spend hours in the lab, and then you will end up with less than a drop in a vial,” Rankin says with a broad, encouraging smile.
Students eagerly file into the adjoining lab, find spots at one of the six large black lab tables, don gloves, and lean down over tubes of DNA. Thumbs pump pipettes as liquids are precisely measured and released into vials. Rankin works the room to inspect progress, lend a gloved hand or answer questions. The students gravitate to the incubator and thermocycler (a device that raises and lowers the temperature of DNA samples in preprogrammed steps) as they work through the 10-step procedure. The first pair finishes at 10:25. Twelve minutes later, everyone is back in room 307.
“Your DNA is getting snipped up!” Rankin says before she launches into instructions for the second procedure, which uses a specialized magnetic rack to separate the DNA from the liquid used to suspend and clean the fragments. “Remember to pipette slowly!” she says as the students stream back to the lab. At 11:45, two boys high-five as they complete the 20-step procedure.

By 2 p.m., everyone has finished the third procedure — amplification of the fragments, the most sensitive of the protocols, which involved a friendly scrum at the centrifuge as nine students waited for their 30-second turn, and lots of excitement at the thermocycler (“It’s the big moment!” one boy announced) — and they are nearing the end of the final 33-step procedure: purifying the DNA libraries.
The energy in the room is palpable.
At 2:09, Ellie Griffin ’21 and Audrey Choi ’20 are the first to place their small vial, carefully, into the carrier that will ultimately hold 12 precious drops of liquid. “Did you mark your vial correctly?” Rankin asks repeatedly as pairs approach, aware that mislabeling could wreak havoc with the data. Within minutes, the carrier is full. The wet lab is over.
Why genome sequencing?
In 2012, Exeter and Stanford University initiated a collaboration dubbed “StanEx,” which brought real-world genetics research into the Exeter curriculum. StanEx was originated by Dr. Seung Kim ’81, Stanford professor of developmental biology and of medicine, along with Rankin and Science Instructor Townley Chisholm. The first StanEx course, which continues today, teaches Exonians how to genetically modify and breed new stocks of fruit flies. To create the stocks, students insert into fruit flies an artificial transposable element (a small chunk of DNA that can change its position within the genome) that carries a “switch” to highlight proteins in fluorescent green, allowing researchers to visually track specific genes in a fly’s development. At Kim’s Stanford lab, scientists use these stocks to research pancreatic disease and diabetes. The stocks, all derived from the StanEx-1 base line, are also made available to researchers around the world.
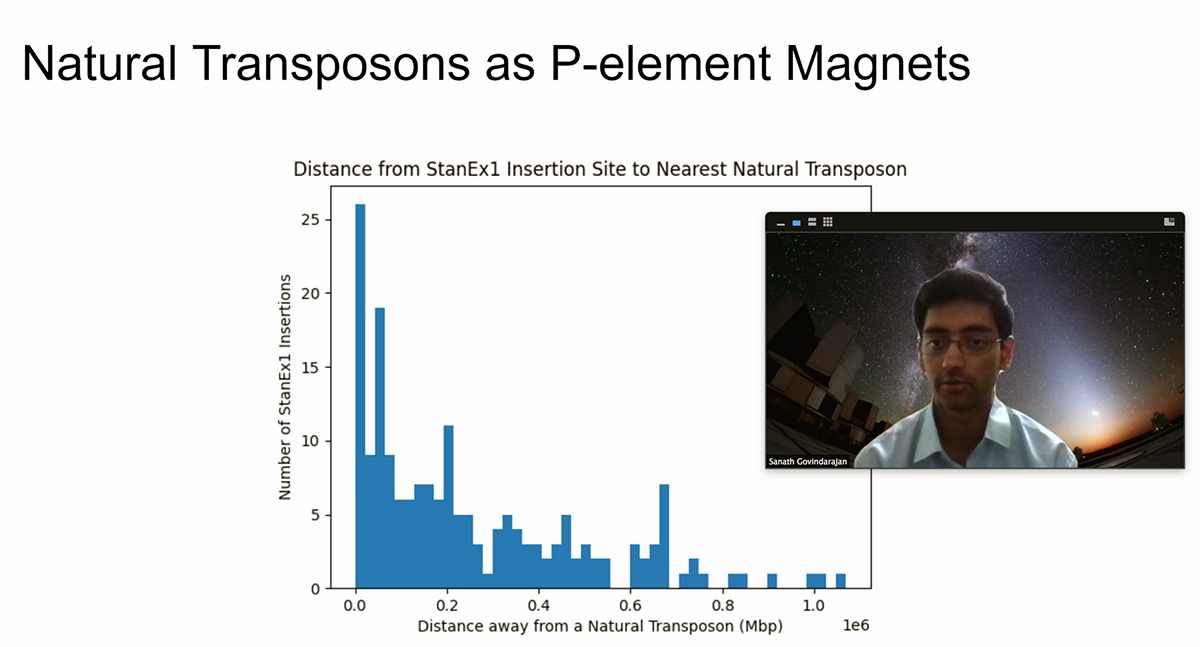
In 2019, a group of almost 90 Exonians, assisted by three science instructors — Rankin, Chisholm and Erik Janicki — along with Kim and Dr. Lutz Kockel from the Stanford lab, published an academic paper in which they addressed a perceived anomaly in the Exeter-created stocks. The process of inserting the transposable element was meant to support totally random locations, but analysis indicated a “hot spot.”
“The fact that we were getting all these things in the same spot was, first of all, a disappointment because you don’t need something in the same spot over and over again. But then it became just plain interesting,” Rankin says. “We were observing that there was this little piece of DNA that was calling our piece to it, and basically they were switching places. It was a really, really cool outcome. I mean, that’s real science.” The question was: Why is this happening?
The Stanford lab tested a few hypotheses, to no avail. “So then comes the next logical question: ‘What else is in the DNA structure that we didn’t know?’” Rankin explains. “We tried to buy something clean, that didn’t have funny background stuff that was going to act like DNA magnets. And that’s where the whole genome sequencing project comes in, because with sequencing, you get all of the DNA. You can look at it and see, ‘Well, what is actually in there?’”

Teacher as learner
You could say Rankin’s route to Exeter dates to the 1950s when H. Hamilton Bissell ’29 offered a newsboy scholarship to her father, Dr. Kenneth Rankin ’59, then a newspaper delivery boy in Cleveland. She and all her siblings in turn attended Exeter. In 1999, at a crossroads, Rankin applied for a teaching job at Exeter and to her top-choice doctoral program. She was accepted by both and decided to teach at Exeter to “get it out of my system.” She’s still working on that.
“You never get bored in a Harkness class because you never quite know what the students are going to ask,” she says. “And you never know how things are going to unfold in front of you.”
To observe Rankin in the classroom is to see many roles: teacher, researcher, scientist and learner. “I think kids respond super well to me when I invest everything I have into learning something new and share with them the places I see for more growth in myself,” she says. “I try to model how to learn rather than present myself as someone who has learned.”
She jokes easily with the students (“You are good at many things,” she told one student during the wet lab, “but slow pipetting is not one of them!”) while making it clear that the work in the classroom is important and of much greater value than the topic at hand. “By becoming learners shoulder-to-shoulder with our students, we authentically model the characteristics we hope to instill — humility, empathy, inquisitiveness, openness, adaptability and continual growth,” Rankin wrote a few years ago in a note to herself that she titled, “Paradigm shift on the concept of teacher.” As one of the founding faculty members of the StanEx project, she has helped it expand over the years into more classes at Exeter, summer internships at the Stanford lab for Exeter students, collaborations with other high schools, and the whole genome sequencing project. All of this has helped feed Rankin’s hunger for “a culture of continual exploration and experimentation.”